A1: Physiological proteomics of Staphylococcus aureus - from stress physiology to in vivo proteomics
[ Project completed ]
The main goal of the project is to use functional genomics technologies, particularly proteomics, to gain new information on the cellular physiology and virulence of S. aureus.
The project is based on the progress made in the first funding period, particularly on the identification and quantification of the entire proteome. The dynamics of the protein inventory, including the protein quality-control system in response to infection-related stimuli, the role of crucial regulons in metabolism and pathogenicity, and some aspects of in vivo proteomics (infection-related conditions) will be analyzed.
Contact
Prof. Dr. Michael Hecker
Institut für Mikrobiologie
Ernst-Moritz-Arndt-Universität Greifswald
Friedrich-Ludwig-Jahn-Straße 15
D-17487 Greifswald
Phone +49 3834 864233
Fax +49 3834 864202
E-Mail: hecker@uni-greifswald.de
Dr. Susanne Engelmann
Institut für Mikrobiologie
Ernst-Moritz-Arndt-Universität Greifswald
Friedrich-Ludwig-Jahn-Straße 15
D-17487 Greifswald
Phone +49 3834 864227
Fax +49 3834 864202
E-Mail: Susanne.Engelmann@uni-greifswald.de
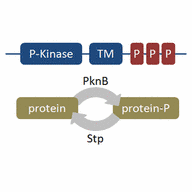
- The domain structure of proteins with PASTA domains (penicillin-binding protein and serine/threonine kinase associated) domains
A - PSTKs, e.g. PknB Mycobacterium tuberculosis
B - Type I PBPs, e.g. PonA Mycobacterium tuberculosis
C - Type II PBPs e.g. SP0336 Streptococcus pneumonia
D - Not characterized e.g. TM1716 Thermotoga maritime
E - Not characterized e.g. TM1716 Thermotoga maritime
ESTPKs have between one and four PASTA repeats.
TM: transmembrane domain
S: signal sequence
P: PASTA-domain
A2: Phosphoproteomic analysis of Staphylococcus aureus: Functional characterization of kinases and identification of their substrates
Reversible protein phosphorylation is a common regulatory mechanism in all living organisms, from bacteria to humans. In this project, the function of the eukaryotic-like serine/threonine kinase (ESTK) PknB and its corresponding phosphatase Stp in Staphylococcus aureus is studied.
In particular, we are interested in signals activating PknB/Stp activity, signal transduction processes, and their role in physiology, virulence, and antibiotic resistance.
Contact
PD Dr. Knut Ohlsen
Institut für Molekulare Infektionsbiologie
Julius-Maximilians-Universität Würzburg
Josef-Schneider-Str. 2, Bau D15
D-97080 Würzburg
Phone +49 931 3182155
Fax +49 931 3182578
E-Mail: knut.ohlsen@mail.uni-wuerzburg.de

A3: Impact of membrane potential on Staphylococcus aureus biofilms
The research topics of A3 have always been centered on staphylococcal physiology and biofilm survival strategies. In the first funding period, the focus was on the physiology of Staphylococcus aureus biofilm; in the second period, it was on generating a membrane potential and regulating nitrate respiration; in the third period, the focus will be on a novel membrane potential-generating system in S. aureus and its impact on biofilm physiology.
Particular attention will be paid to “physiological hotspots” in S. aureus biofilms resulting from spatiotemporal changes in expression of genes of energy metabolism.
Contact
Prof. Dr. Friedrich Götz
LS Mikrobielle Genetik
Eberhard-Karls-Universität Tübingen
Auf der Morgenstelle 28
D-72076 Tübingen
Phone +49 7071 2974635 or -36
Fax +49 7071 295937
E-Mail: friedrich.goetz@uni-tuebingen.de
Prof. Dr. Katharina Riedel
Institut für Mikrobiologie
Ernst-Moritz-Arndt-Universität Greifswald
Friedrich-Ludwig-Jahn-Strße 15,
D-17487 Greifswald
Phone: +49 3834 86 4200
E-Mail: riedela@uni-greifswald.de
A6: X-ray analysis of enzymes and virulence factors of pathogenic staphylococci
We propose to determine three-dimensional structures of selected staphylococcal proteins in order to enhance understanding of their mechanism of function and develop compounds that modulate their function, for example by interfering with ligand binding. Our goals for the next funding period are to build on work performed during the last funding period and provide a structural foundation for the interaction of several staphylococcal proteins with cognate ligands.
The congruency of the proposed research is to establish a foundation for a better understanding of key metabolic and transport processes of staphylococci, and eventually use this information for improved therapy.
Contact
Prof. Dr. Thilo Stehle
Interfakultäres Institut für Biochemie
Eberhard-Karls- Universität Tübingen
Hoppe-Seyler-Str. 4
D-72076 Tübingen
Phone +49 7071 2973043
Fax +49 7071 295565
E-Mail: thilo.stehle@uni-tuebingen.de
A8: A systems biology perspective of regulatory and metabolic adaptation of Staphylococcus aureus to infection-related conditions
Previous work has shown that S. aureus responds rapidly and effectively to infection-relevant stimuli by adaptations of its gene expression and reorganization of protein complexes and metabolic pathways. For a comprehensive understanding of S. aureus physiology and virulence, we will now elucidate adaptation of functional interaction networks with a focus on host challenges by a combination of experimental, bioinformatics and systems biology approaches.
This reference model transcends the idea of static protein complexes by integrating the full dynamics of the regulatory and metabolic networks in S. aureus and will expand existing S. aureus pathogen models.
Contact
Dr. Jan Pané-Farré
Institut für Mikrobiologie
Ernst-Moritz-Arndt-Universität Greifswald
Friedrich-Ludwig-Jahn-Straße 15
D-17487 Greifswald
Phone: 03834-86 4237
E-Mail: janpf@uni-greifswald.de
Prof. Dr. Thomas Dandekar
Lehrstuhl für Bioinformatik, Biocenter
Julius-Maximilians-Universität Würzburg
Am Hubland
D-97074 Würzburg
Phone +49 931 3184551
Fax +49 931 8884552
E-Mail: dandekar@biozentrum.uni-wuerzburg.de
| |||||