Z1: An integrated view on adaptation of Staphylococcus aureus
This project will apply data management, presentation and visualization with the aim of providing a unified S.aureus platform (AureoView) for public accessibility of available data and knowledge.
Project goals will be accomplished by: (i) complementing and extending the AureoWiki, for instance by regulatory information, pathway maps, and gene expression data, (ii) integration of the information from all topic-specific databanks and inclusion of data generated in infection-related settings and (iii) utilization of new visualization techniques for the dissemination of data, e.g., augmented web-based presentation of S.aureus models.
Contact
Prof. Dr. Thomas Dandekar
Lehrstuhl für Bioinformatik, Biozentrum
Julius-Maximilians-Universität Würzburg
Am Hubland
D-97074 Würzburg
Phone +49 931 3184551
Fax +49 931 8884552
E-Mail: dandekar@biozentrum.uni-wuerzburg.de
Dr. Jörg Bernhardt
Institut für Mikrobiologie
Ernst-Moritz-Arndt-Universität Greifswald
Friedrich-Ludwig-Jahn-Str. 15
D-17487 Greifswald
Phone +49 3834 864211
Fax +49 3834 864202
E-Mail: joerg.bernhardt@uni-greifswald.de
Dr. Ulrike Mäder
Interfakultäres Institut für Genetik und Funktionelle Genomforschung
Ernst-Moritz-Arndt-Universität Greifswald, Universitätsmedizin
Friedrich-Ludwig-Jahnstr. 15a
D-17475 Greifswald
Phone: +49 3834-865873
E-mail: ulrike.maeder@uni-greifswald.de
Z2: Proteomics of Staphylococcus aureus
As a central project, Z2 provides an analytical platform for the entire TRR. Consequently, access to sophisticated tools for proteome analysis – including separation of complex protein mixtures and consecutive identification, characterization and quantification of proteome samples – is ensured for all partners.
Furthermore, a robust workflow for a quantitative phosphoprotein analysis will be established. To meet the requirements of high-throughput screening of clinical S. aureus isolates, a workflow for a global, label-free proteome analysis based on SWATH methodology will be established and optimized. Newly developed proteomics methods will be applied in cooperative projects with partners of the CRC-TRR34.
Contact
PD Dr. Dörte Becher
Institut für Mikrobiologie
AG Massenspektrometrie
Ernst-Moritz-Arndt-Universität Greifswald
Friedrich-Ludwig-Jahn-Straße 15
D-17487 Greifswald
Phone +49 3834 864230
Fax +49 3834 864202
E-Mail: dbecher@uni-greifswald.de
Prof. Dr. Katharina Riedel
Institut für Mikrobiologie
Ernst-Moritz-Arndt-Universität Greifswald
Friedrich-Ludwig-Jahn-Straße 15,
D-17487 Greifswald
Phone: +49 3834 86 4200
E-Mail: riedela@uni-greifswald.de
Z3: In vivo imaging of Staphylococcus aureusinfections
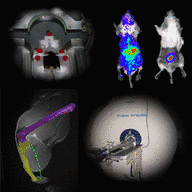
The aim of project Z3 is the implementation of in vivo imaging platform techniques (bioluminescence, fluorescence, magnetic resonance) to visualize the dynamics of Staphylococcus aureus infections and corresponding morphological and physiological changes in host tissues.
High-resolution morphology and functional/physiological parameters will identify systemic or local responses to infection in real-time images, building the basis for the development of novel strategies to prevent staphylococcal diseases. Overall, these data will provide an overview of the dynamics of bacterial spread in the host and its 3D distribution.
Contact
PD Dr. Knut Ohlsen
Institut für Molekulare Infektionsbiologie
Julius-Maximilians-Universität Würzburg
Josef-Schneider-Str. 2, Bau D15
D-97080 Würzburg
Phone +49 931 3182155
Fax +49 931 3182578
E-Mail: knut.ohlsen@mail.uni-wuerzburg.de
Z4: Metabolomics of Staphylococcus aureus
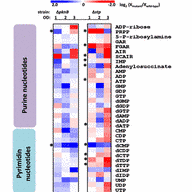
Microbial metabolomics plays a pivotal role in the understanding of the variability and dynamics of bacterial cell metabolism because it measures the direct biochemical response to the environment, i.e., the metabolites. Project Z4 is a central service project in the CRC-TRR34.
The metabolomics platform will serve as a readily available analytical service offering state-of-the-art bioanalytical methods in the CRC-TRR34. In particular, we will provide insights into extracellular and intracellular metabolite compositions using NMR spectroscopy, HPLC-MS, HPLC-MS/MS and GC-MS, GCxGC-TOFMS techniques. With our expertise, we will increase the knowledge about staphylococcal physiology under different cellular physiological states.
Contact
Prof. Dr. Michael Lalk
Ernst-Moritz-Arndt-University Greifswald
Institute of Biochemistry
Felix-Hausdorff-Str. 4
17489 Greifswald
Germany
Telephone: +49-(0)3834-86-4867
Telefax: +49-(0)3834-86-4885
E-Mail: lalk@uni-greifswald.de
| |||||