- Linking N-metabolism and virulence gene expression – a working hypothesis:
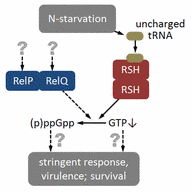
The stringent control is mediated by the (p)ppGpp synthase Rel, which results in the re-programming of gene expression (stable and unstable RNAs) either directly or via lowering of the GTP level. The repressor CodY upon binding of isoleucine and/or GTP represses genes involved in nitrogen metabolism. N-limitation results in de-repression of codY regulated target genes. The proposed regulatory circuits presumably also affect the expression of major virulence genes such as toxins and adhesins.
B1: Role of the stringent response in bacterial survival, antibiotic tolerance and cell death
The stringent stress response is mediated by the messenger molecule (p)ppGpp. For Staphylococcus aureus, we recently showed that the (p)ppGpp synthase activity of the RSH (RelA-SpoT-homologue) enzyme is necessary for the intracellular expression of phenol-soluble modulins (PSMs) and that these molecules are required to the escape from the phagolysome.
Two small (p)ppGpp synthases, RelP and RelQ are part of the cell-wall stimulon and mediate tolerance towards cell-wall active antibiotics. In the future, we will continue to decipher the influence of the stringent response in S. aureus on intracellular survival, persistence and antibiotic tolerance.
Contact
Prof. Dr. Christiane Wolz
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Eberhard-Karls-Universität Tübingen
Elfriede-Aulhorn-Straße 6
D-72076 Tübingen
Phone +49 7071 2980187
Fax +49 7071 295165
E-Mail: christiane.wolz@med.uni-tuebingen.de
B4: Regulation of methionine metabolism in Staphylococci: Impact on fitness and virulence
In this project we will analyze the regulatory factors, proteins and metabolites involved in S. aureus methionine biosynthesis control under varying physiological conditions
We will study the dynamics of met locus transcription as well as the stability of enzymes and metabolites of the methionine pathway, and investigate the impact of both the lack of methionine biosynthesis or its overproduction on the fitness, virulence and pathogenesis of S. aureus. Finally, we will unravel the structural prerequisites and biological significance of the met-T-box riboswitch interaction with initiator tRNAifMet.
Contact
PD Dr. Wilma Ziebuhr
Institut für Molekulare Infektionsbiologie
Julius-Maximilians-Universiät Würzburg
Josef-Schneider-Str. 2
D-97080 Würzburg
Phone +49 931 3182155
Fax +49 931 3182578
E-Mail: w.ziebuhr@mail.uni-wuerzburg.de
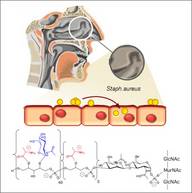
B5: Wall teichoic acid structure variation in Staphylococcus-host interaction
The wall teichoic acid (WTA) polymers of S. aureus interact with several human binding partners and play crucial roles in colonization and infection. We found that WTA structure varies in different S. aureus clonal lineages with respect to backbone structure and glycosylation with α- or β-linked GlcNAc and that these variations affect interaction with host factors and transducing bacteriophages.
We propose to explore the structural variation of WTA in vivo and consequences for the ability of S. aureus to colonize, activate or evade immune recognition, or exchange mobile genetic elements.
Contact
Prof. Dr. Andras Peschel
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Zelluläre und Molekulare Mikrobiologie
Eberhard-Karls-Universität Tübingen
Elfriede-Aulhorn-Str. 6
D-72076 Tübingen
Phone +49 7071 2981515
Fax +49 7071 293435
E-Mail: andreas.peschel@uni-tuebingen.de
| |||||